Dataset Analysis#
Prior to executing training tasks, performing comprehensive dataset feature analysis is essential. This process enables systematic evaluation of training memory requirements through statistical characteristics (e.g., edge distribution in individual graph structures), informs parameter configuration strategies (including neural network irreducible representations), and guides performance expectation assessments while establishing model evaluation baselines.
DFT Data Features#
Command Line Interface#
The dock analyze dataset features command provides comprehensive analysis of DFT dataset characteristics, enabling users to make informed decisions about model architecture and training strategy.
dock analyze dataset features [OPTIONS] DATA_PATH
Basic Usage#
# Analyze dataset with default settings
dock analyze dataset features /path/to/your/data
# Analyze dataset with specified orbital types
dock analyze dataset features . --common-orbital-types s3p2d1f1
# Analyze dataset considering parity and with parallel processing
dock analyze dataset features /path/to/data --consider-parity -p 4
Options Explained#
Option |
Alias |
Description |
Default |
|---|---|---|---|
|
|
Number of parallel processes for analysis. |
|
|
|
Directory tier level for DFT data organization: |
|
|
|
User-defined common orbital types specification (e.g., |
Auto-detected |
|
Include parity consideration in irreps analysis |
|
|
|
|
Display help message |
N/A |
Output Interpretation#
The command generates a comprehensive analysis report with multiple sections:
Example Output for Water (H\(_2\)O) Dataset#
📊 BASIC DATASET INFO
-----------------------
• Spinful: False
• Parity consideration: False
• Total data points: 4,999
🧪 ELEMENT & ORBITAL INFO
---------------------------
• Elements included: H, O (2 elements)
• Orbital source: auto_detected
• Common orbital types: s3p2d1f1
🎯 IRREPS INFORMATION
-----------------------
Irreps Type Irreps Dimension
.................... .................................................. ..........
Common orbital 15x0e+24x1e+22x2e+18x3e+8x4e+3x5e+1x6e 441
Suggested 16x0e+24x1e+24x2e+24x3e+8x4e+4x5e+2x6e 518
Exp2 32x0e+16x1e+8x2e+4x3e+2x4e+2x5e+2x6e 214
Trivial 32x0e+32x1e+32x2e+32x3e+32x4e+32x5e+32x6e 1568
💡 RECOMMENDATIONS
--------------------
1. Moderate dataset size - regular training recommended
2. High-dimensional irreps - consider dimensionality reduction techniques
Key Information Sections#
1. Basic Dataset Info
Spinful: Indicates if the system is spin-polarized (True) or spin-restricted (False)
Parity consideration: Whether spatial inversion symmetry is considered in irreps
Total data points: Number of DFT calculation samples available for training
2. Element & Orbital Info
Elements included: Chemical elements present in the dataset with count
Orbital source: Whether orbital types were auto-detected or user-specified
Common orbital types: Orbital basis specification in
sNpNdNfNformatExample:
s3p2d1f1means 3 s-orbitals, 2 p-orbitals, 1 d-orbital, 1 f-orbital
3. Irreps Information The irreps (irreducible representations) table shows four different symmetry-adapted representations:
Irreps Type |
Purpose |
When to Use |
|---|---|---|
Common orbital |
Symmetry-adapted basis from detected orbital types |
Default choice for most models |
Suggested |
Optimized representation based on symmetry analysis |
For best performance with the current dataset |
Exp2 |
Reduced-dimensional representation |
When computational efficiency is prioritized |
Trivial |
Complete (non-symmetry-adapted) representation |
As a baseline or for debugging |
The notation NxMe indicates N occurrences of irreducible representation with angular momentum M and parity e (even). For example, 15x0e means 15 scalar (ℓ=0) even-parity representations.
4. Dimension Analysis Each irreps type has a total dimension indicating the feature vector size. This directly impacts:
Model parameter count
Memory requirements
Training time
Model capacity
5. Recommendations Context-specific suggestions based on dataset characteristics:
Dataset size guidance: Small (<100 samples), Moderate (100-6000), Large (>6000)
Architecture suggestions: Based on irreps dimensionality and system properties
Training strategies: Recommendations for regularization, augmentation, etc.
Practical Usage Scenarios#
1. Initial Dataset Assessment#
# Quick initial assessment with defaults
dock analyze dataset features /your/data/inputs
Use this to get a baseline understanding of your dataset’s scale and complexity.
2. Orbital Basis Optimization#
# Compare different orbital basis choices
dock analyze dataset features ./inputs --common-orbital-types s10p5d3
dock analyze dataset features ./inputs --common-orbital-types s12p7d5f3
Compare dimension reductions and select the optimal balance between accuracy and efficiency.
3. Training Configuration Planning#
Based on the output:
Total data points: Determines train/validation/test split ratios
Irreps dimensions: Guides model architecture design and hidden layer sizes
Memory estimate: Informs GPU memory requirements and batch size selection
4. Performance Benchmarking#
# Analyze with parity consideration for comparison
dock analyze dataset features /path --consider-parity
Compare dimensions with and without parity to understand the impact of symmetry constraints.
Interpreting Dimension Values#
Common orbital dimension: 441
Represents the symmetry-adapted feature space size
Determines the input layer size of the neural network
Higher values capture more complex chemical environments but require more parameters
Suggested vs Common:
The Suggested Irreps are usually the recommended choice.
Suggested dimension (518) > Common (441): The algorithm suggests expanding the representation
This indicates potential benefits from a richer feature space
Exp2 dimension: 214
Approximately half of common orbital dimension
Useful for testing if reduced features still capture essential physics
Trivial dimension: 1568
Non-symmetry-adapted, complete basis
Much larger dimension highlights the compression achieved by symmetry adaptation
Advanced Analysis Tips#
Tier Number Selection: Use
-toption to match your directory structure# For data organized as: # - data/dft/tire1/material1 # - data/dft/tire1/material2 # - data/dft/tire2/material3 # - ... dock analyze dataset features data -t 1
Parallel Processing: For large datasets, adjust parallel processing
# Use 8 cores for faster analysis dock analyze dataset features /path -p 8
Orbital Type Notation: The orbital type string follows
sNpNdNfNformat where:s3: 3 s-orbitals (typically 1s, 2s, 3s)p2: 2 p-orbitals (typically 2p, 3p)d1: 1 d-orbital (typically 3d)f1: 1 f-orbital (typically 4f)
Common Output Patterns#
Dataset Type |
Expected Irreps Dimension |
Typical Recommendations |
|---|---|---|
Small molecule |
2000-5000 |
Data augmentation, transfer learning |
Bulk material |
200-600+ |
High-capacity model, regularization |
Spin-polarized |
Approximately 2× larger |
Ensure spin-symmetry handling |
Multi-element |
Higher dimension |
Element-specific embeddings |
Next Steps After Analysis#
Model Architecture Design: Use the irreps dimensions to design your neural network layers
Training Configuration: Set batch size based on memory estimates
Performance Expectations: Use dataset size to estimate training convergence time
Validation Strategy: Split data appropriately based on total samples
This analysis provides the foundational understanding needed to configure effective machine learning models for materials science applications, ensuring appropriate model capacity for your specific dataset while maintaining computational efficiency.
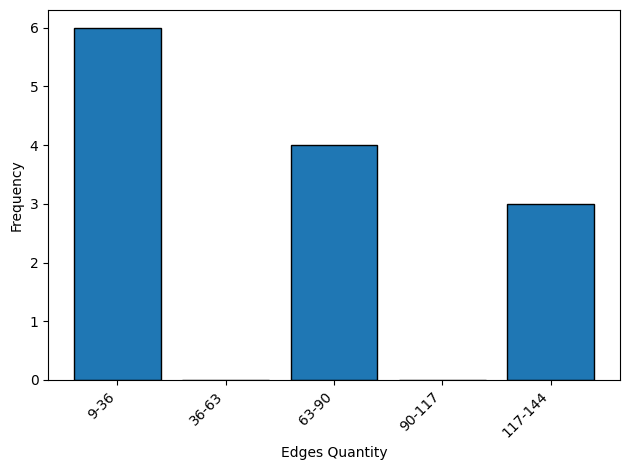
Edge number statistics#
User can use the terminal tool to analyze the edge number of the structures in the dataset. The edge number is related to the memory usage and speed of the training/inference processes.
dock analyze dataset edge ./inputs -p 4
The description of the arguments can be shown by
dock analyze dataset edge -h
Usage: dock analyze dataset edge [OPTIONS] DATA_PATH
Statistic and show edge related information.
Options:
-p, --parallel-num INTEGER The parallel processing number, -1 for using all of the cores. [default: -1]
-t, --tier-num INTEGER The tier number of the dft data directory, -1 for <data_path>/dft, 0 for <data_path>/dft/<data_dirs>, 1 for <data_path>/dft/<tier1>/<data_dirs>, etc. [default: 0]
--edge-bins INTEGER The bins count width, default is auto decided.
--plot-dpi INTEGER The bin count plot DPI. [default: 300]
-h, --help Show this message and exit.
For advanced users, deeph-dock provides the DatasetAnalyzer class.
from deepx_dock.analyze.dataset.analyze_dataset import DatasetAnalyzer
data_path = "./inputs"
parallel_num = 4
tier_num = 0
edge_bins = None
plot_dpi = 300
inspector = DatasetAnalyzer(
data_path=data_path,
n_jobs=parallel_num,
n_tier=tier_num,
)
features = inspector.gen_dft_features()
inspector.statistic_edge_quantity(
features=features,
bins=edge_bins,
)
[do] Locate all DFT data directories ...
[rawdata] Processing DFT data in inputs/dft.
+-[search]: 13it [00:00, 10429.60it/s]
[rawdata] Found 13 structures in inputs/dft.
inspector.plot_edge_quantity(dpi=plot_dpi)
Dataset split generation#
User can use the terminal tool to split the dataset into train set, validation set, and test set. A json file containing the split structure names is dumped.
dock analyze dataset split ./inputs -p 4
cat ./dataset_split.json
{"train": ["25001", "1001", "4992", "25000", "4990", "4991", "4995"], "validate": ["25003", "4994"], "test": ["1002", "4993", "25002", "1000"]}
The description of the arguments can be shown by
dock analyze dataset split -h
Usage: dock analyze dataset split [OPTIONS] DATA_PATH
Generate train, validate, and test data split json file.
Options:
-p, --parallel-num INTEGER The parallel processing number, -1 for using all of the cores. [default: -1]
-t, --tier-num INTEGER The tier number of the dft data directory, -1 for <data_path>/dft, 0 for <data_path>/dft/<data_dirs>, 1 for <data_path>/dft/<tier1>/<data_dirs>, etc. [default: 0]
--split-ratio <FLOAT FLOAT FLOAT>...
The train_ratio, validate_ratio and test_ratio of the split data. [default: 0.6, 0.2, 0.2]
--split-max-edge-num INTEGER The max edge number of the split data, -1 for no constraint. [default: -1]
--split-rng-seed INTEGER The random seed for processing dataset. [default: 137]
-h, --help Show this message and exit.
The class API is also provided.
from deepx_dock.analyze.dataset.analyze_dataset import DatasetAnalyzer
data_path = "./inputs"
parallel_num = 4
tier_num = 0
split_ratio = [0.6, 0.2, 0.2]
split_max_edge_num = -1
split_rng_seed = 137
inspector = DatasetAnalyzer(
data_path=data_path,
n_jobs=parallel_num,
n_tier=tier_num,
)
features = inspector.gen_dft_features()
inspector.generate_data_split_json(
features=features,
train_ratio=split_ratio[0],
val_ratio=split_ratio[1],
test_ratio=split_ratio[2],
max_edge_num=split_max_edge_num,
rng_seed=split_rng_seed,
)
[do] Locate all DFT data directories ...
[rawdata] Processing DFT data in `inputs/dft`.
+-[search]: 13it [00:00, 10233.85it/s]
[rawdata] Found `13` structures in `inputs/dft`.
Data Split: 100%|██████████| 13/13 [00:00<00:00, 27.47it/s]
[info] Total available data dirs: 13
[info] Data split json saved to ./dataset_split.json.